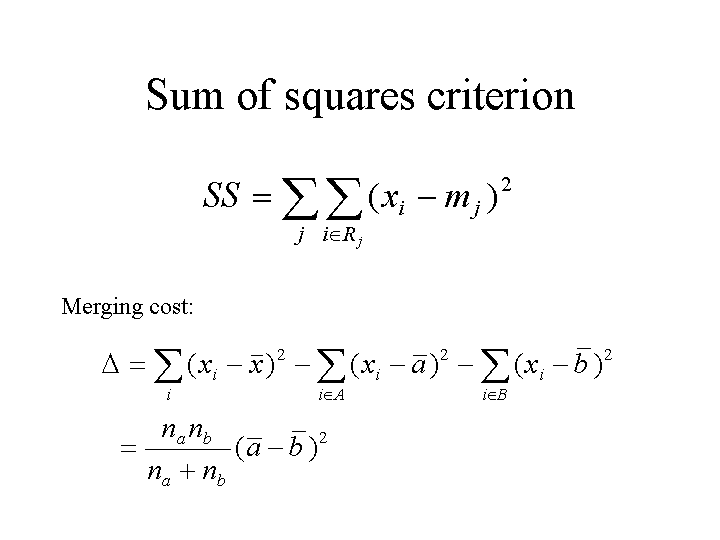
After considering a few examples, it becomes clear that separation alone is not a sufficient criterion for clustering. Separation reveals itself as valleys in the histogram. If the dataset is uniformly distributed among its range, there are no valleys and thus no separation. But it wouldn't be right to put everything into one cluster, one abstraction bin, since that would not "preserve distinctions." The dataset could be composed of two very different subgroups, whose density along this variable happens to be uniform. In the uniform case, the natural binning is an equally spaced one. It preserves the distinction between distant points but not between nearby points. We can explain this as a desire for balance as well as separation. There are many choices for how to trade-off balance and separation. Today we discuss one way.
The choices we have when clustering can be broken down as follows:
A criterion which achieves a nice trade-off of balance and separation is the sum-of-squares criterion. The evaluate a given clustering, we compute the mean of the data in each cluster, then add up the sum of squared differences between each point and its cluster mean. Note that this is just the variance of the cluster times the size of the cluster, summed over all clusters. We want to minimize the criterion, i.e. a large sum of squares is bad. This criterion clearly wants clusters to be compact. It also wants balance, because big clusters cost more than small ones with the same variance.
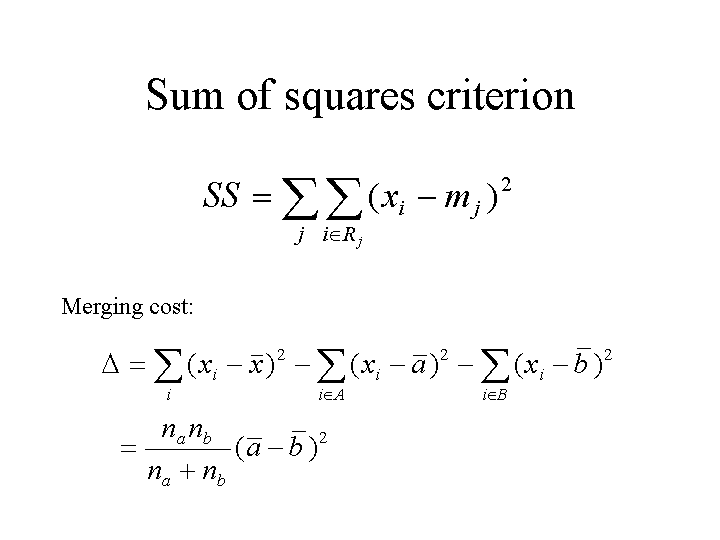
A disadvantage of k-means algorithms is that the answers they give are not unique. While there may be only one clustering with minimum sum of squares, these algorithms will not necessarily find it every time. Iterative re-assignment is a local search algorithm: it makes small changes to the solution and sees if the objective got better. Local search is also called hill-climbing, by analogy to an impatient hiker who tries to find the highest peak by always walking uphill. This strategy can stop at local minima, where the criterion is not optimized but no improvement is possible by making small changes. Which local minimum you stop at depends on where you were when you started. Hence the k-means algorithms give different results depending on how they make their initial guess. A typical initial guess is to pick k data points at random to be the means.
clus1.r
clus1.s
Class examples
Two routines are provided for getting a set of bin breaks according to one of the above schemes. The function break.hclust(x,n) uses Ward's method to divide the range of x into n bins. It plots the breaks and returns a break vector, just like bhist.merge. It also gives a trace of the cost of each merge. This allows you to choose the number of clusters. An interesting number of clusters is one which directly precedes a sudden jump in merging cost.
The function break.kmeans(x,n) does the same thing but with k-means. Note that break.kmeans does not always give the same answer every time you run it.
If you want a full hierarchy, you can use the built-in function hclust. It returns a hierarchy object that you can view as a tree using plot in R or plclust in S. You can also view it as a nested set of bin breaks using plot.hclust.breaks. See the example code.