Resources for the Unlimited Sensing Framework
Contents
If you use this code, please acknowledge the following papers:
- Sampling Theorem On Unlimited Sampling Bhandari et al. in Proc. of Sampling Theory and Applications Conference (SampTA), 2017. • (DOI)
- Main Paper On Unlimited Sampling and Reconstruction Bhandari et al. in IEEE Trans. on Signal Proc., 2020. • (DOI)
- First Hardware Paper Unlimited Sampling from Theory to Practice: Fourier-Prony Recovery and Prototype ADC Bhandari et al. in IEEE Trans. on Signal Proc., 2021. • (DOI)
Additionally, US Patent can be found here
% Code for Demo. % Unlimited Sampling of Bandlimited Functions. % Ayush Bhandari (ayush@alum.MIT.edu) % The code is confidential and for collaborative purposes only. clc close all clear all set(0,'defaultlinelinewidth',2)
Overview Comment
This codes uses a moderate amount of noise added to the samples.
Empirically, this is well handled by the amplitude enfocing constraint on the residue function.
Occasionally, the reconstruction may fail.
Basic Function Definitions
% Following functions handles are defined that are useful later. % * Fractional Part % * Origin Centered Modulo Function % * Rounding Function for Enforcing Amplitude Constraints % * Pi-Bandlimited Kernel % * Difference Order based on Dynamic Range and Sampling Rate e = exp(1); % Fractional Part frac = @(x) x-floor(x); % Origin Centered Modulo Function RD = @(x,L) round(x./(2*L)).*2*L; % Round Function M = @(f,T) 2*T.*( frac(f./(2.*T) + 0.5) -0.5 ); % Bandlimited Kernel K = @(x) sinc(x); % Difference Order DO_US = @(T,L,Bg) ceil((log(L) - log(Bg))/log(T*pi*exp(1))); % Threshold Line LT = @(L,ts) L.*ones(numel(ts),1); % Phase-unwrapping PU = @(data,L) unwrap(data.*(pi./L))./(pi/L);
Prepare Analog and Digital Signals
% Time t = -12:0.005:20; t = t(:); %Ts = 1/(2*pi*e)-0.001; Ts = 0.005/0.055; %Ts = 0.05/0.005; Td = ceil(1/Ts); locs = 1:Td:numel(t); ts = t(locs); % Amplitudes of Random Bandlimited Signal Z = rand(numel(t),1) + 1i.*rand(numel(t),1); F = fft(K(t)).*Z; f = real(ifft(F)); f = f./max(abs(f)); f = f-mean(f); %f = f + 0.3.*t;
Modulo-Folded Samples
% Conventional Samples data = f(locs); data0 = data; % Modulo Threshold L = randi([1 10],1,1)/100; L = 0.01 + (0.1-0.01).*rand(1,1); L = 1/60; % Modulo Samples mdata = M(f,L); % Modulo Samples mdm = M(data,L);
Reconstruction from Folded Samples
% Maximum Derivative DO = DO_US(mean(diff(ts)),L,max(f)); % Reconstruct Initial Data rec = M(diff(mdm,DO),L); tsDO = ts(1:end-DO); plot(tsDO,rec,tsDO,LT(L,tsDO),tsDO,-LT(L,tsDO)); % Reconstruction Loop clear Kn ds res al bl ds = diff(mdm,DO); res = rec(:) - ds(:); res = RD(res,L); cntr = cell(DO);
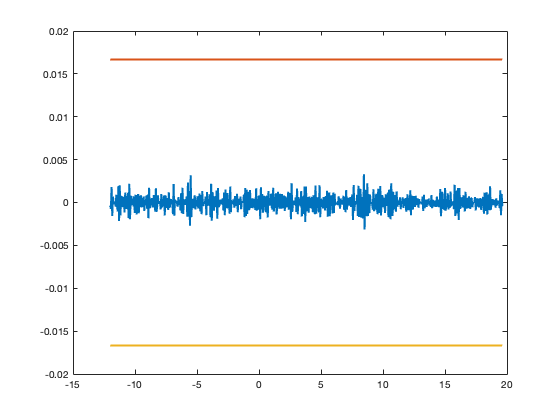
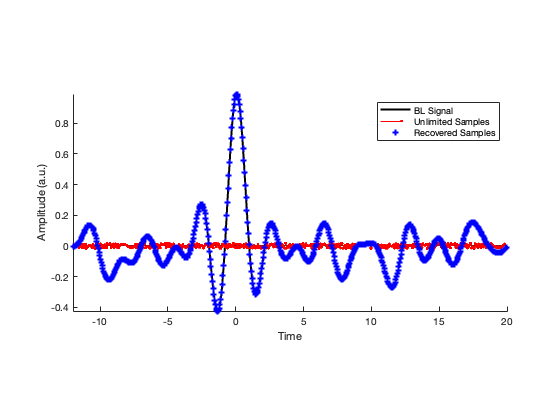
clear S2 Kn_iter Rec_Sig al bl egN cntr al = res; % Initialized Variables bl = res; % Initialized Variables egN = al; % Initialized Variables g0 = ceil(max(abs(data))); Bg = ceil(g0/(2*L))*2*L; % Ceiling of Max-norm J = ceil(6*Bg/L); % Empirically This works Well % Loop Backwards for Reconstruction for k = 1: DO-1; cntr{k} = egN; S2 = cumsum([0 ;(cumsum([0;egN]))]); Kn_iter = floor((-S2(J+1) + S2(1))/(12*Bg) + 0.5) bl = cumsum([0; bl]); bl = RD(bl,L); bl = bl + 2*Kn_iter*L; egN = bl; end close all Rec_Sig = cumsum([0; bl])+mdm(:); offset = (data(locs(1)) - Rec_Sig(1))/L; Rec_Sig = Rec_Sig + offset*L; %Rec_Sig = Rec_Sig - mean(Rec_Sig); hold on; plot(t,f,'k') stem(ts,mdm,'r.','MarkerSize',4) plot(ts,Rec_Sig,'b*','MarkerSize',2) axis tight xlabel('Time') ylabel('Amplitude (a.u.)') legend('BL Signal','Unlimited Samples','Recovered Samples') pbaspect([4 2 2])
Kn_iter =
0
Kn_iter =
0
Kn_iter =
0
Kn_iter =
0
Kn_iter =
0
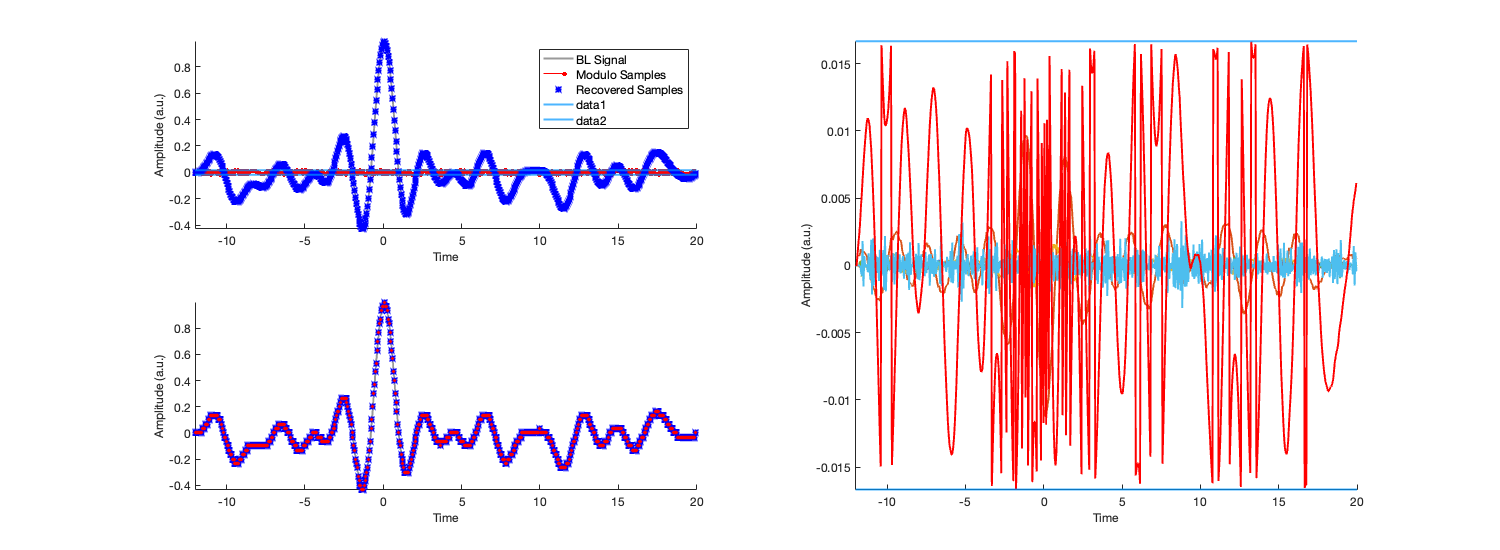
subplot(2,2,1) hold on; plot(t,f,'Color',[1 1 1].*0.6) stem(ts,mdm,'Marker','.','MarkerSize',10,'MarkerFaceColor',[1 0 0],... 'LineWidth',0.5,... 'Color',[1 0 0]) RP = plot(ts,Rec_Sig,'b*','MarkerSize',3); legend('BL Signal','Modulo Samples','Recovered Samples') h1 = plot(t,-LT(L,t)); h2 = plot(t,+LT(L,t)); axis tight h1.Color = [0 150 255]./255; h2.Color = [0 150 255]./255; h1.Color(4) = 0.7; h2.Color(4) = 0.7; RP.Color(4) = 0.2; xlabel('Time') ylabel('Amplitude (a.u.)') %offset*L*0 subplot(2,2,3) hold on; stairs(t,f - M(f,L),'Color',[1 1 1].*0.6,'LineWidth',2) h1 = plot(ts,cumsum([0 ; bl])+offset*L,'b*'); h1.Color(4) = 0.3; plot(ts, data - mdm,'rsq','MarkerSize',2) axis tight xlim([min(t) max(t)]) xlabel('Time') ylabel('Amplitude (a.u.)') subplot(2,2,[2 4]) hold on; for DO_iter = 1:DO MDO(DO_iter,:) = [zeros(DO_iter,1);M(diff(mdm,DO_iter),L)]; end plot(ts,MDO,ts,[0 ; M(diff(M(data,L)),L)],'r') h1 = plot(t,-LT(L,t)); h2 = plot(t,+LT(L,t)); axis tight h1.Color = [0 150 255]./255; h2.Color = [0 150 255]./255; h1.Color(4) = 0.7; h2.Color(4) = 0.7; axis tight xlim([min(t) max(t)]) xlabel('Time') ylabel('Amplitude (a.u.)') FS = 12; X = 600; Y = 220; fig = gcf; set(fig, 'Position', [0, 0, X, Y]); fig = gcf; set(findall(fig,'-property','FontSize'),'FontSize',FS) set(fig, 'Position', [0, 0, X, Y].*2.5)
Report on Reconstruction
clc disp(sprintf(['Noise Power in dB: %g'], SNR_db)); disp(sprintf(['Max-norm (Bg): %g'], Bg)); disp(sprintf(['ADC Threshold: %g'], L)); disp(sprintf(['Maximum Derivative Order: %g'], DO)); disp(sprintf(['Reconstruction MSE is %g'], immse(Rec_Sig,data)));
Unrecognized function or variable 'SNR_db'. Error in Unlimited_Sampling_Demo (line 270) disp(sprintf(['Noise Power in dB: %g'], SNR_db));